Synthesis center for cell biology
We are putting together a proposal to make a Synthesis Center for the field of cell and molecular biology. Its goal would be to synthesize the vast quantities of available cell and molecular data (protein types, locations, abundances, interactions) into both conceptual and quantitative models, that allow us to explain and predict the remarkable transition from nonliving molecules to living cells. The center (if they choose our proposal) would be funded by the National Science Foundation for 5-10 years and be housed at the Allen Institute for Cell Science in Seattle. During this workshop, I'd love to bounce around ideas for the synthesis center, and to identify points of intersection between this proposed center and your favorite tool or area.
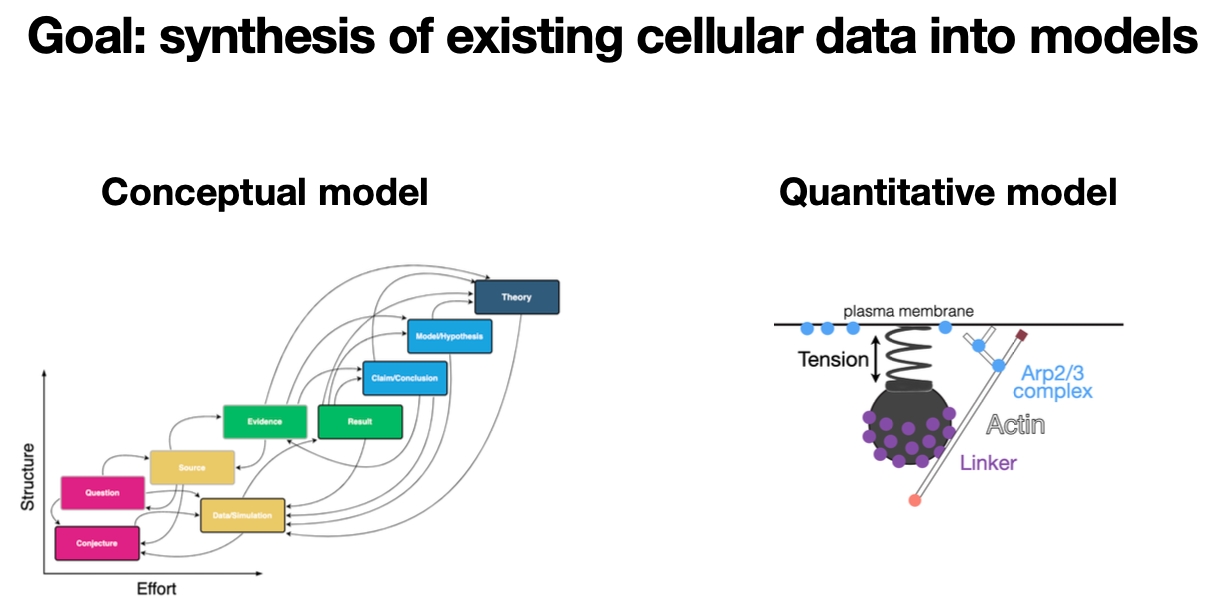
While most of the "big data" cell biology community is focused on creating new data sets, we are proposing to create conceptual and quantitative models from existing data. To make conceptual models, I'm proposing that we use the power of the discourse graph schema to structure the state of knowledge for our favorite research question(s). Furthermore, we'll extend the discourse graph schema to guide our ongoing research contributions to address these questions. We call these results graphs.