Translate Logseq Knowledge Graph to Systems Biology Network Diagrams
| Translate Logseq Knowledge Graph to Systems Biology Network Diagrams | |
|---|---|
| Homepage | |
| Description | Looking at possibilities to generalized knowledge graph in Logseq based on Discourse Graph developed by Prof. Joel Chan and David Vargas for knowledge synthesis, with the possibility to export the information to Systems biology graphical notation markup language to create the network/graph.
|
| Associated Publications | 10.1515/jib-2020-0016, 10.1016/j.tplants.2010.03.005, 10.1002/psp4.12155 |
| Contributors | Akila Wijerathna-Yapa |
We are interested in adopting a modified version of Prof. Joel Chan, and David Varga's discourse graph Roam extension to literature synthesis for our ongoing text-mining project for biological network modeling. Having a knowledge graph in Logseq with its essential sections - Hypothesis, Evidence, and Experimental Results will enable us (researchers) to get insights into knowledge synthesis/biological network modeling.
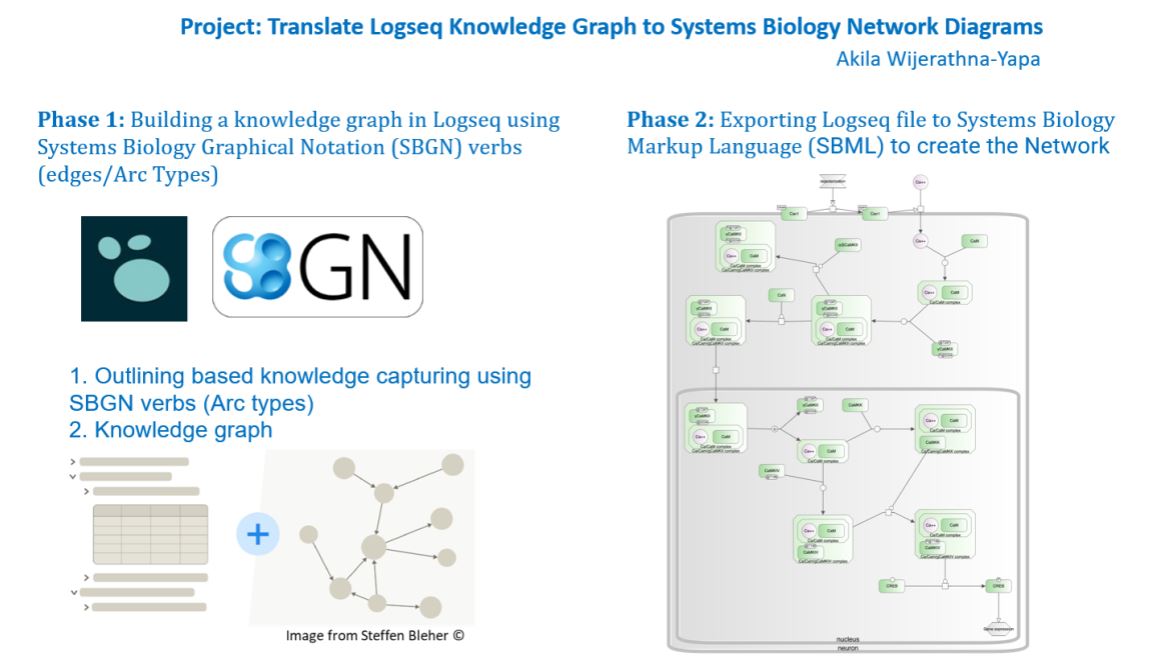
For the first phase, we look opportunity to create a knowledge graph in Logseq using limited discourse graph features — Hypothesis, Evidence, and Experimental Results, using standardized Bio-ontologies. This will offer a method for formalizing biological knowledge, such as that pertaining to genes, anatomy, and phenotypes, in complicated hierarchies made of concepts and rules. For this we can probably use Systems Biology Graphical Notations (SBGN) based arcs — i.e. SBGN Activity Flow language.
In the second phase, we intend to convert the knowledge network into System Biology Graphical representation using SBGN with an expectation to import the SBGN Biological network into Systems Biology Markup Language (SBML) for downstream computational/mathematical simulations.
The Challenge: Building Knowledge netwok in Logseq and translate into SBGN.
s