Synthesis center for cell biology: Difference between revisions
(Created page with "We are putting together a proposal to make a Synthesis Center for the field of cell and molecular biology. Its goal would be to synthesize the vast quantities of available cell and molecular data (protein types, locations, abundances, interactions) into both conceptual and quantitative models, that allow us to explain and predict the remarkable transition from nonliving molecules to living cells. The center (if they choose our proposal) would be funded by the National Sc...") |
(Creating a synthesis center to enable grassroots contributions for conceptual and quantitative models in cell biology.) |
||
| Line 1: | Line 1: | ||
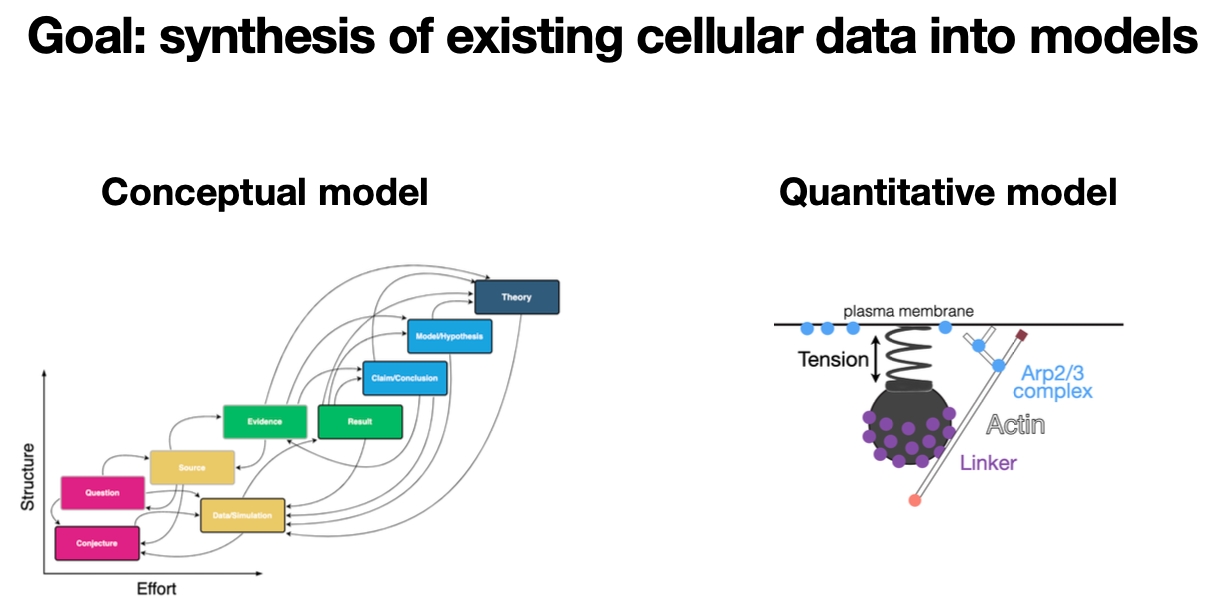
We are putting together a proposal to make a Synthesis Center for the field of cell and molecular biology. Its goal would be to synthesize the vast quantities of available cell and molecular data (protein types, locations, abundances, interactions) into both conceptual and quantitative models, that allow us to explain and predict the remarkable transition from nonliving molecules to living cells. The center (if they choose our proposal) would be funded by the National Science Foundation for 5-10 years and be housed at the Allen Institute for Cell Science in Seattle. During this workshop, I'd love to bounce around ideas for the synthesis center, and to identify points of intersection between this proposed center and your favorite tool or area. | We are putting together a proposal to make a Synthesis Center for the field of cell and molecular biology. Its goal would be to synthesize the vast quantities of available cell and molecular data (protein types, locations, abundances, interactions) into both conceptual and quantitative models, that allow us to explain and predict the remarkable transition from nonliving molecules to living cells. The center (if they choose our proposal) would be funded by the National Science Foundation for 5-10 years and be housed at the Allen Institute for Cell Science in Seattle. During this workshop, I'd love to bounce around ideas for the synthesis center, and to identify points of intersection between this proposed center and your favorite tool or area. | ||
[[File:Conceptual and quantitative models.png| | [[File:Conceptual and quantitative models.png|center|500x500px]] | ||
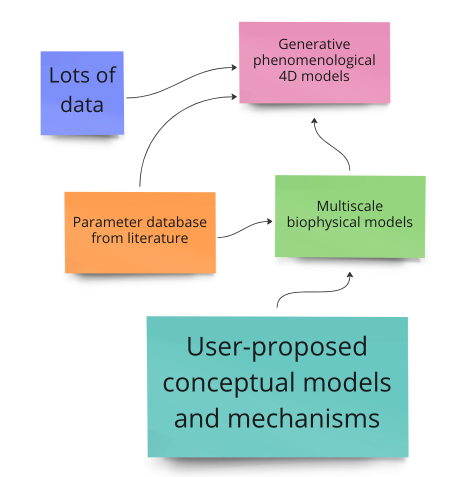
While most of the "big data" cell biology community is focused on creating new data sets, we are proposing to | While most of the "big data" cell biology community is focused on creating new data sets, we are proposing to synthesize existing data into quantitative and conceptual models. For one set of quantitative models, we are using large datasets of the locations and interactions of cellular components to train generative (a la DALL-E-2) models of cells. The goal is for these synthetic cells to behave realistically in novel environments. For those models to be predictive, we need to constrain them with 1) quantitative parameter values from the literature and 2) mechanistic and biophysical information about the underlying processes. We need some help with #1 (an NLP challenge). For #2, we are building a platform to make mechanistic biophysical models of cellular processes that are interoperable, modular, and accessible. But how do we as a field synthesize existing cell biology data into higher-level concepts, models, and theories? | ||
[[File:Purpose and users of cell biology discourse graphs.png| | [[File:Quantitative model generation.png|center|350x350px]] | ||
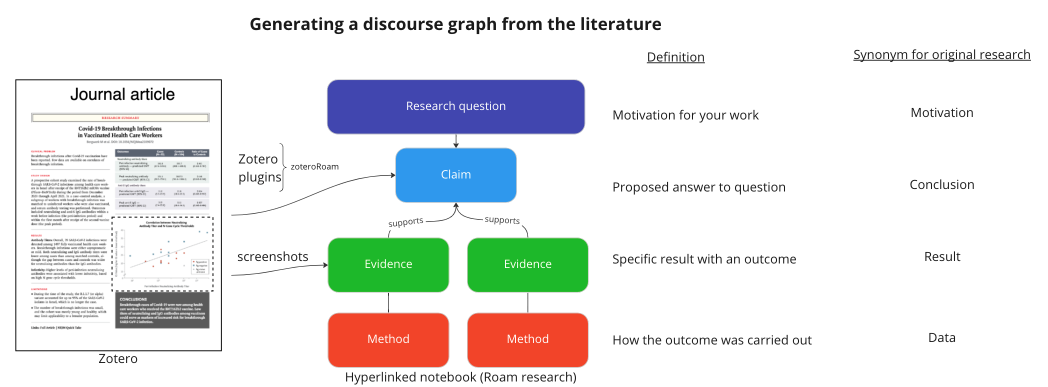
To make conceptual models, we would like to use the power and modularity of the discourse graph schema - Questions, Claims, and Evidence - to structure the state of knowledge for our favorite research question(s). Furthermore, we'll extend the discourse graph schema to guide our ''ongoing'' research contributions to address these questions. We call these [https://youtu.be/P0KUt2yrUkw results graphs]. Our lab has begun to create discourse and results graphs to track our understanding and contributions to our current research questions. Using Roam Research and Joel Chan's discourse graph extension, we classify a given research Question, collect Evidence from the literature and our lab notebooks, and use them to support Conclusions, which claim to address the research question. It is early days, but this schema appears to help students structure their thinking, track their progress, and - most importantly - frame their work less as an individual endeavor and more as a contribution to a collective project (i.e. we are all trying to uncover the answer together). | |||
[[File:Purpose and users of cell biology discourse graphs.png|center|350x350px]] | |||
[[File:Schematic_of_discourse_graphs.png|center|700x700px|Schematic of a discourse graph generated from the literature, and the analogous terms for ongoing research.]] | |||
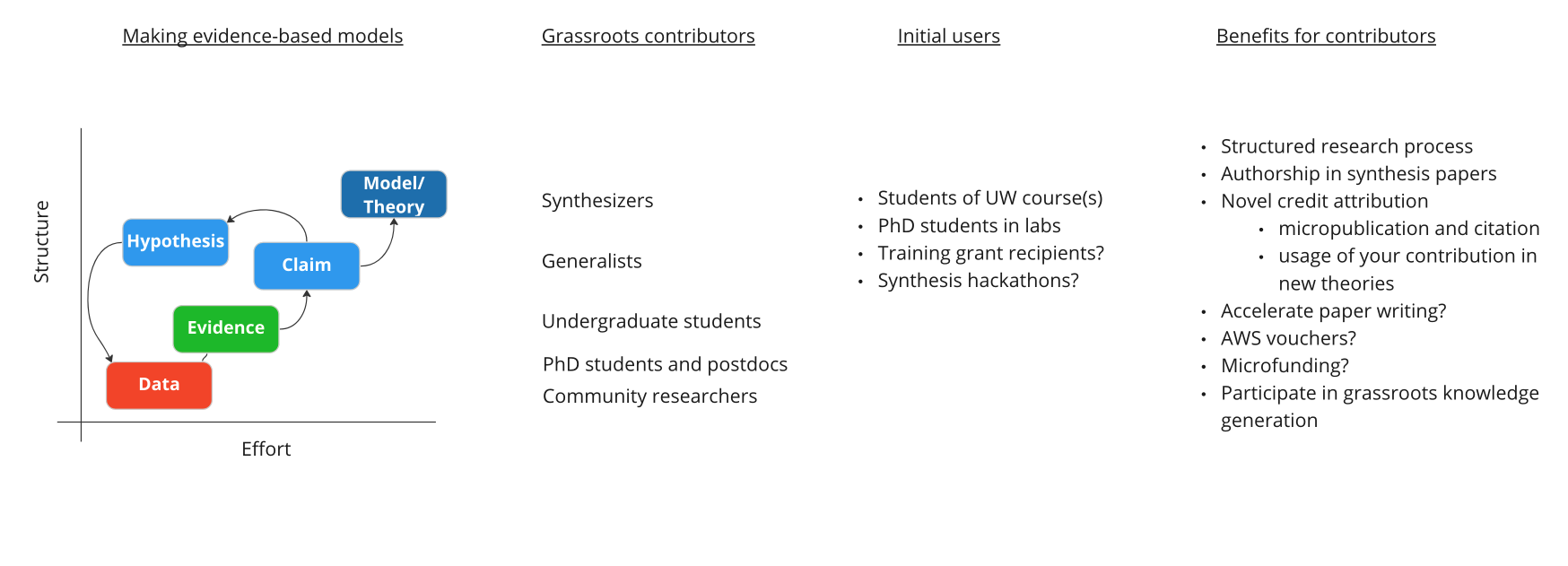
With the current tooling, paired with some ease-of-use improvements, and a 'captive audience' in the form of initial users who are also beneficiaries of the synthesis center, we think that discourse and results graphs in cell biology will allow for ''grassroots'' contributions from students, scientists, and community researchers, to build overarching concepts, models, and theories in cell biology. | |||
[[File:Progress to theories.png|center|800x800px]] | |||
Lots of questions about this proposed center! | |||
* What are the major roadblocks for adoption by the cell biology community? What ease-of-use improvements will tip the balance of benefits vs overhead for using these discourse graph tools? | |||
* What is the role of cartoon models in building our conceptual models, and can we interoperate between/merge the cartoons (a la knowledge graphs, and [https://prior.allenai.org/projects/diagram-understanding computer vision])? Or at the very least use them as the visual backdrop for our discourse graphs? | |||
* In practice, what is the relationship between a conceptual model and a quantitative model? Do we need to formalize components in a knowledge graph? Or just make the information (models/claims with related evidence and arguments) available and accessible to quantitative modelers? | |||
* What does a minimum discourse graph micropublication platform look like? Does it involve Obsidian Publish? What are the minimum features (versioning)? | |||
* What is the best way to interoperate between labs' (and researchers') discourse graphs? Federated? Centralized platform with branches and mergers? | |||
* Can we assist users with extracting evidence and claims from the literature, with an NLP tool? Can our 'captive audience' of students and researchers provide the necessary training data? | |||
* Can we use NLP to help convert between a discourse graph and drafting a narrative (i.e. paper or proposal)? Is the structure of a manuscript sufficiently formulaic to pull this off (I think yes)? | |||
* Can we use NLP to help to extract all the instances of a given parameter value from the literature? | |||
Revision as of 10:18, 10 November 2022
We are putting together a proposal to make a Synthesis Center for the field of cell and molecular biology. Its goal would be to synthesize the vast quantities of available cell and molecular data (protein types, locations, abundances, interactions) into both conceptual and quantitative models, that allow us to explain and predict the remarkable transition from nonliving molecules to living cells. The center (if they choose our proposal) would be funded by the National Science Foundation for 5-10 years and be housed at the Allen Institute for Cell Science in Seattle. During this workshop, I'd love to bounce around ideas for the synthesis center, and to identify points of intersection between this proposed center and your favorite tool or area.
While most of the "big data" cell biology community is focused on creating new data sets, we are proposing to synthesize existing data into quantitative and conceptual models. For one set of quantitative models, we are using large datasets of the locations and interactions of cellular components to train generative (a la DALL-E-2) models of cells. The goal is for these synthetic cells to behave realistically in novel environments. For those models to be predictive, we need to constrain them with 1) quantitative parameter values from the literature and 2) mechanistic and biophysical information about the underlying processes. We need some help with #1 (an NLP challenge). For #2, we are building a platform to make mechanistic biophysical models of cellular processes that are interoperable, modular, and accessible. But how do we as a field synthesize existing cell biology data into higher-level concepts, models, and theories?
To make conceptual models, we would like to use the power and modularity of the discourse graph schema - Questions, Claims, and Evidence - to structure the state of knowledge for our favorite research question(s). Furthermore, we'll extend the discourse graph schema to guide our ongoing research contributions to address these questions. We call these results graphs. Our lab has begun to create discourse and results graphs to track our understanding and contributions to our current research questions. Using Roam Research and Joel Chan's discourse graph extension, we classify a given research Question, collect Evidence from the literature and our lab notebooks, and use them to support Conclusions, which claim to address the research question. It is early days, but this schema appears to help students structure their thinking, track their progress, and - most importantly - frame their work less as an individual endeavor and more as a contribution to a collective project (i.e. we are all trying to uncover the answer together).
With the current tooling, paired with some ease-of-use improvements, and a 'captive audience' in the form of initial users who are also beneficiaries of the synthesis center, we think that discourse and results graphs in cell biology will allow for grassroots contributions from students, scientists, and community researchers, to build overarching concepts, models, and theories in cell biology.
Lots of questions about this proposed center!
- What are the major roadblocks for adoption by the cell biology community? What ease-of-use improvements will tip the balance of benefits vs overhead for using these discourse graph tools?
- What is the role of cartoon models in building our conceptual models, and can we interoperate between/merge the cartoons (a la knowledge graphs, and computer vision)? Or at the very least use them as the visual backdrop for our discourse graphs?
- In practice, what is the relationship between a conceptual model and a quantitative model? Do we need to formalize components in a knowledge graph? Or just make the information (models/claims with related evidence and arguments) available and accessible to quantitative modelers?
- What does a minimum discourse graph micropublication platform look like? Does it involve Obsidian Publish? What are the minimum features (versioning)?
- What is the best way to interoperate between labs' (and researchers') discourse graphs? Federated? Centralized platform with branches and mergers?
- Can we assist users with extracting evidence and claims from the literature, with an NLP tool? Can our 'captive audience' of students and researchers provide the necessary training data?
- Can we use NLP to help convert between a discourse graph and drafting a narrative (i.e. paper or proposal)? Is the structure of a manuscript sufficiently formulaic to pull this off (I think yes)?
- Can we use NLP to help to extract all the instances of a given parameter value from the literature?